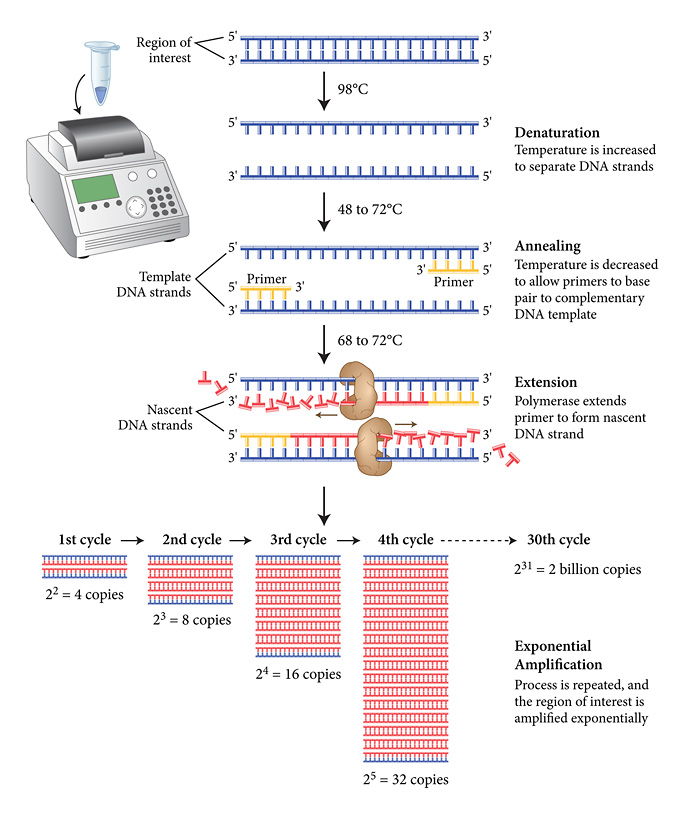
First, the DNA of the desired (extracted) gene is added to an extraction mix, and incubated at 95 degrees Celsius, in which the DNA is denatured, that is, the double helix stricture is broken apart into 2 free-floating strands, releasing the DNA into the liquid.
Next, we add dilution mix, and pipet our sample up and down repeatedly to create a homogeneous solution, with an equal concentration of solute throughout.
Then centrifuge your sample for 2 minutes, separating the DNA, promoters, primers, and liquid within the sample. Mix with red PCR mix, which may contain the appropriate promoters and DNA polymerase.
Place into the PCR machine, which hats up (denaturing) and cools down (annealing) the DNA, to amplify it, or produce many copies that can be studied.
The sample is then put left to sit at room temperature, in which the DNA is annealed, or brought back together through primers that create complementary strands, by base pairing with the promoter to create a complementary DNA template. At this point, only promoters and primers have bonded to the DNA
The sample is the extended, when DNA polymerase extends from the primer to form a new DNA strand, complementary to the free-floating strands of DNA. These strands can bond, and the process begins again, creating new strands.
https://www.neb.com/~/media/NebUs/Page%20Images/Applications/DNA%20Amplification%20and%20PCR/pcr.jpg
No comments:
Post a Comment